Recently I started work at Curoverse, which I like to describe as “the cloud for genomes,” and I like to describe my job as “being paid to work on open-source bioinformatics software.”
I think the official site has some more informative words to say on this subject and talks about the vision of enabling biomedical information sharing to improve research and clinical outcomes or something. Anyway, I’m an intern so thankfully I haven’t had to pitch the idea repeatedly every day until I can present it in a concise and powerful manner 🙂
I’ve been having a great time. I mean, it’s (a) open-source (b) bioinformatics (c) python (d) has nothing at all to do with cruel cruel robots. 😛
I’ve learned a lot about genomes, sequencing, and all the pitfalls of genomes, aka why Arvados (the open-source platform Curoverse is working on) is needed:
That is fifty phased genomes, shipped on multi-terabyte hard drives to us. Sneakermaillll!
Here are some important things I have learned during the month or so I’ve been at the job so far:
1) Our mascot is called Dax. It is adorableeeeee

No, we don’t have plushies of Dax yet, but soon!
Oh! I could easily revolve this and make a 3d model that I could print off this weekend. Hmm. *adds to todo list*
2) In software, open-source is much more established and can be seen as a boon by customers, especially by researchers and clinical or governmental infrastructure.
3) Having a stable source of income is great! Having really knowledgeable co-workers is great! Having an office with an ergonomic chair and free tea is great! Plus, all my work is open-source, so I can easily refer to it in the future, or point people / my friends at it.
4) It’s been super interesting to see the sales and development cycle. I’ve been observing Curoverse’s strategies for finding the so-called “product-market fit” and the continual effort to find and work on what customers want — in some senses, selling something before developing it.
Okay, I’m sort of just rambling here, so I’ll talk a little bit about the environment I’ve found myself in and the work I’ve been doing.
1) Environment:
The location is great. It’s a few minutes walk from the South Station stop on the Red Line, and within walking distance to Chinatown as well as views of the water.
It’s all open-style desks, no cubicles. Everyone’s right next to each other, and furthermore EVERYONE IS ON IRC. (even some of our customers apparently!). It works surprisingly well, since I’ve had cases where I’ll ask someone a question (in the team chat) and someone else will give me a faster way to do it / enable me to do it myself. It’s still not zephyr with its classes that let you sort through a multi-threaded conversation easily, but it’s nice.
Unlike what I’ve come to expect of software development, a lot of the folks are older / married / have kids, and also have more engineering experience of course. There’s also a GPL ninja floating around…
What really puzzled me at first was calling one section of software developers “engineering” and another “science,” but it makes sense now. I just expected the engineering team to be building more robots I guess. ^^;
The engineering team operates on “agile” development (or lean?) — anyway the main thing to me is they are always talking about stories, which amuses me. I imagine them typing away drafting a really epic novel sometimes.
The science team, which I have found myself on, has standups every day as well, were we all stand up and present to the rest of the team what we’ve been up to. Standing up hasn’t seemed very effective at keeping our standup short though o_o
Every Friday there is also free lunch err I mean, a company meeting, which keeps everyone on the same page.
Oh! I got to attend the first Curoverse social.
Delicious pizza (flatbread) and bowling! It was candlepin bowling, where the pins are flattened like candlesticks instead of like the traditional bowling pin and the balls are solid and a bit bigger than a softball.
(yep, people are wearing their work outfits, which vary a lot).
2) What I’ve been working on.
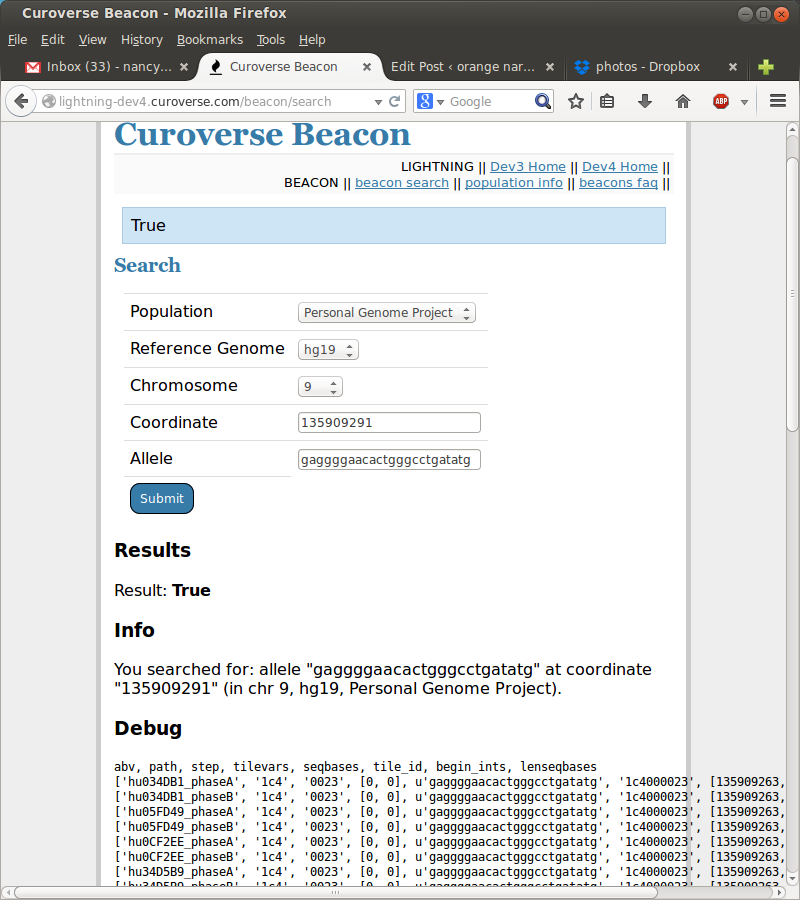
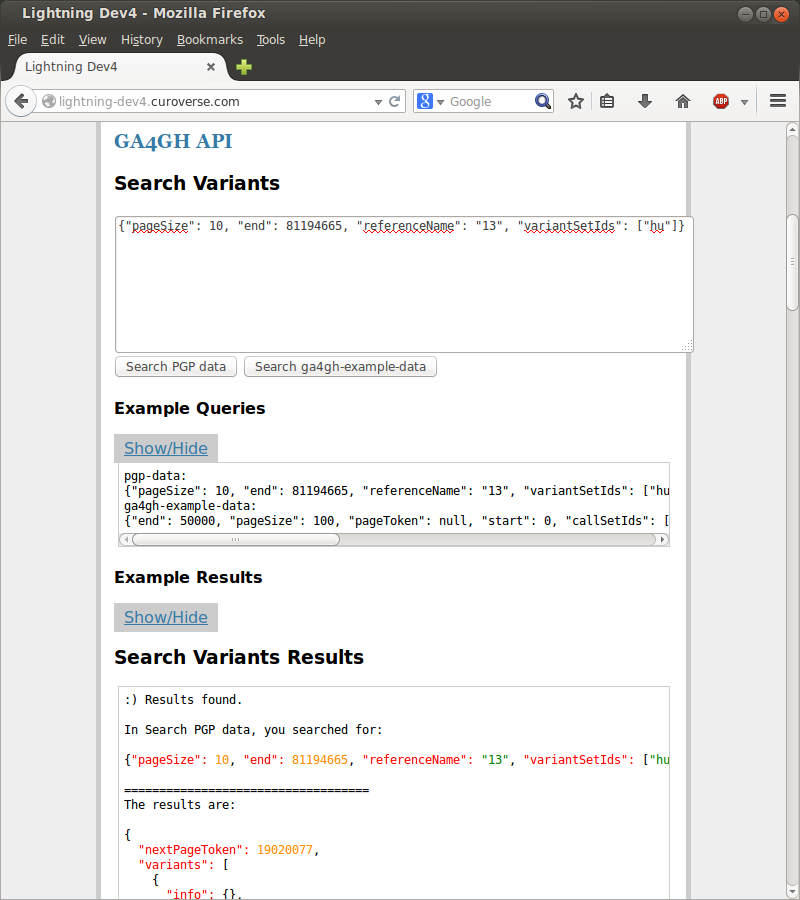
Right away I got involved in working with GA4GH, aka the Global Alliance for Genomics and Health, or more accurately working on projects the GA4GH groups determine are important. I made a little python+flask webapp and learned how to serve it using uWSGI and nginx,as well as learned a bit of working with postgresql (such as the fact that DISTINCTs take a significant chunk of time, aka seconds, because they query across the whole table!).
I’ve also been playing around with javascript to make POSTs to an API and return the results.
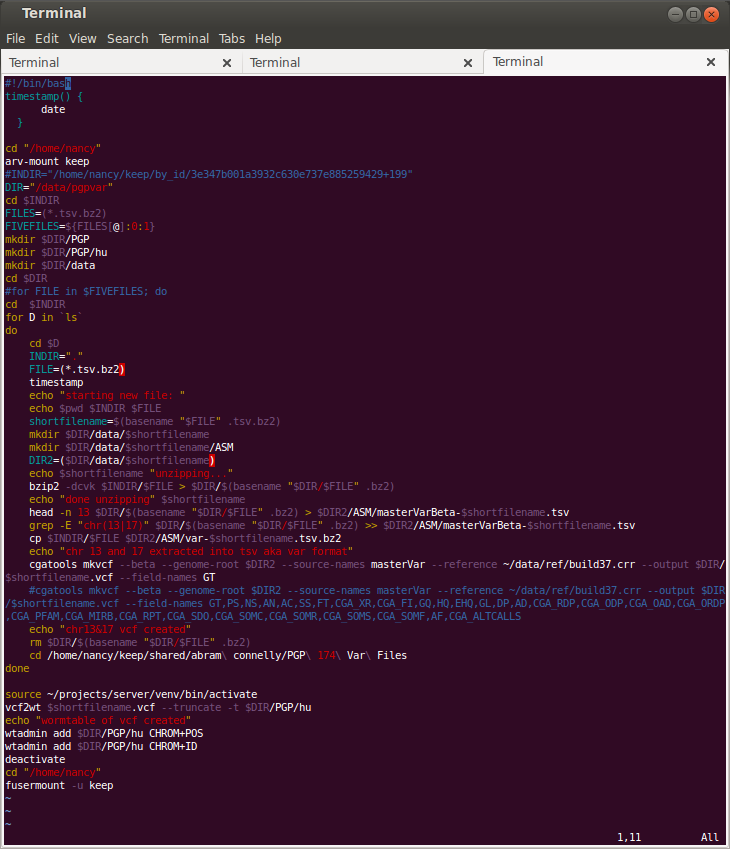
OH! AND WRITING TERRIBLE BASH SCRIPTS because apparently bioinformaticians not only can’t agree on whether to 0-index or 1-index, but also make these convoluted file formats followed by convoluted file-format-conversion tools that require specifically named files in certain directories all documented in a PDF somewhere on the internet. WHYYYY
Hmm, well, now I feel like I haven’t done much since I can sum it all up in three images. Regardless, I sure have learned a lot while getting paid to do so, and I’ve always had plenty of work to do. There’s in fact a GA4GH conference this weekend where my supervisor might demo some of my work as part of the science team’s work, which is nifty.
See you all next time!