I needed to create some FASTA files of important regions of the human genome for work. BRCA1 & 2 are important in breast cancer research, SMA is implicated in spinal muscular atrophy, and KIR and MHC are important for your immune system (e.g. why you have to have organ donor “compatibility”). I wrote a bash script that used Entrez Direct to automatically download these files from the NCBI servers.
FASTA files
If you just want the FASTA files to play with, they are here.
They were downloaded from the NCBI website and based on the NCBI Gene database coordinates against hg38.
source code
https://github.com/nouyang-curoverse/GA4GH_regions
This repository contains a link to download the final FASTA files, the ids.csv file I used as the master list of “mhc” and “kir” genes, the bash script file, and the xsl “xml transform” file I used to extract the information I needed from the xml file.
what i learned
Hey, what’s a gene anyway? If you use various databases (ensembl, ncbi, genenames, lumc, omim, lrg), you”ll get a whole range of coordinates for the same gene.
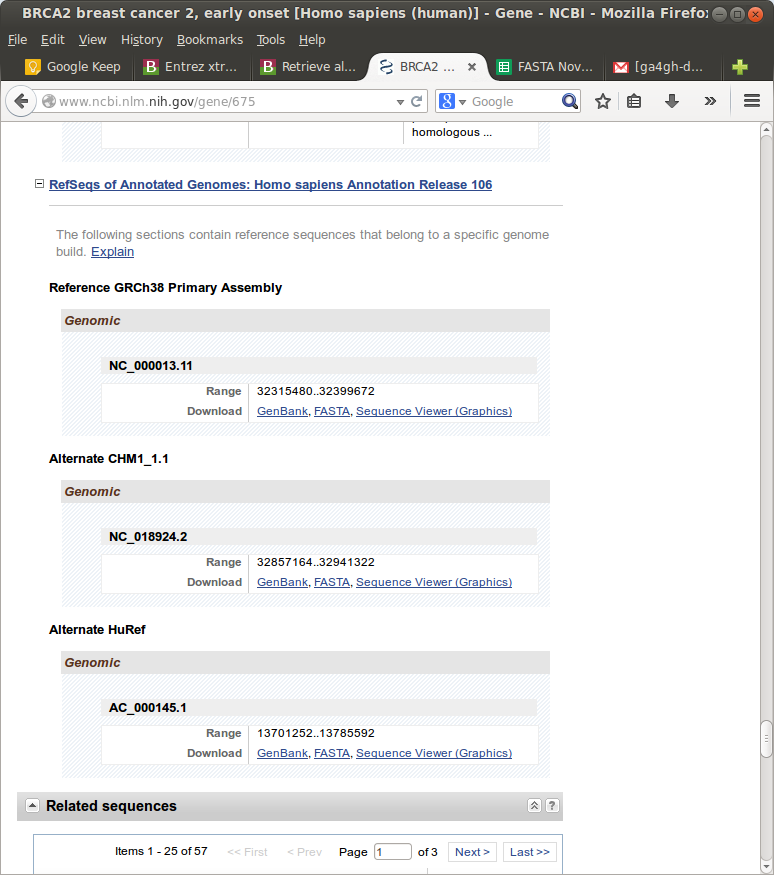
Take, for instance, BRCA2:
| Start | End | Length | Source |
| 32,889,611 | 32,974,403 | 84,792 | ensembl |
| 32,884,617 | 32,975,809 | 91,192 | lrg |
| 32,889,617 | 32,973,809 | 84,192 | ncbi entrez |
| 32,889,616 | 32,973,808 | 84,192 | OMIM |
| 32,890,598 | 32,972,907 | 82,309 | COSMIC sanger |
| 32,889,641 | 32,907,422 | 17,781 | CGAP |
In the end I standardized around using the NCBI Gene database and ignored the rest.
Hey, what’s the KIR gene region anyway? Turns out there’s a gazillion KIR genes, and there’s not exactly a “list” of them. Same for HLA. I just used my best human judgement and culled them from searches on NCBI Gene. From the git repo Readme:
For the HLA genes, I used the IMGT list of gene names, found at ftp://ftp.ebi.ac.uk/pub/databases/ipd/imgt/hla/fasta/ . For the KIR genes, I used NCBI Gene query to list all the KIR genes. http://www.ncbi.nlm.nih.gov/gene and
"Homo sapiens"[porgn] AND KIR
The final list is in ids.csv.
The manual process was to download the FASTA file from the NCBI Gene database.
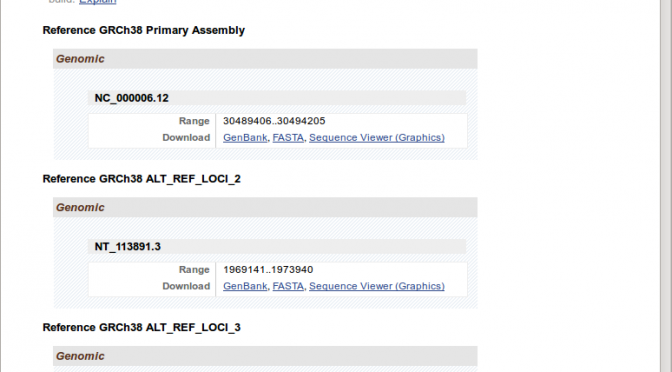
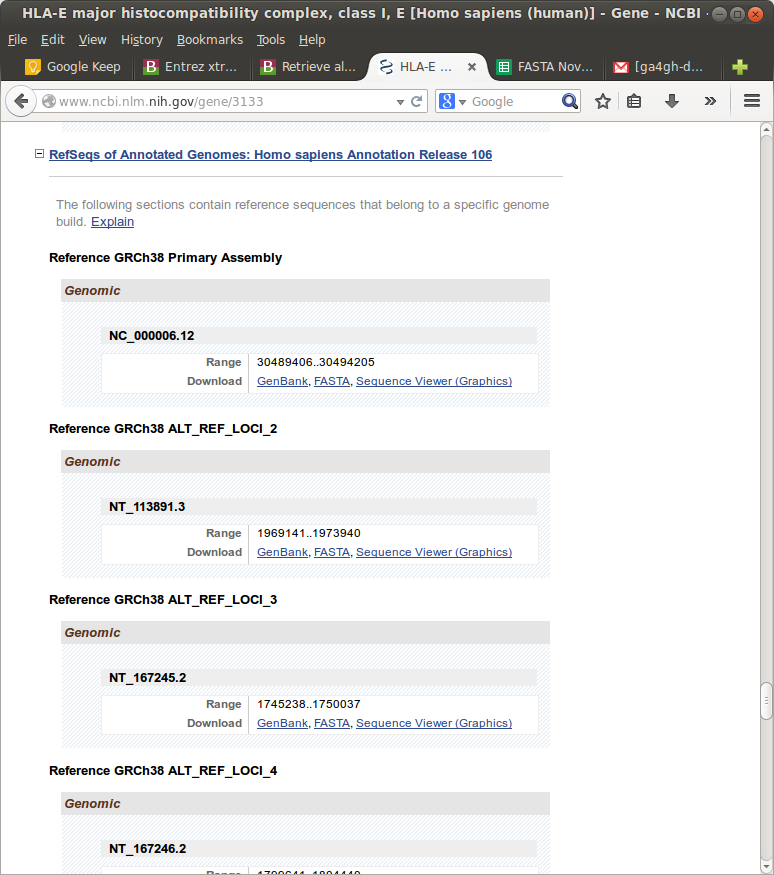
But for some genes, that is the KIR and HLA genes, each gene (e.g. just HLA-E) has many “alternate locii” versions.
Downloading these by hand would take days. Therefore, I needed to figure out a way to script this. My initial thought was to write a scraper using Google Sheet Scripts or a python library like Beautiful Soup. However, I thought this was dumb, because this NCBI Gene site is clearly the front-end to some database that hopefully had an API for programmatic access of some kind.
After a few days (seriously, this all took way longer than I thought it would) and with help from the biostars community I was able to figure out how to use Entrez Direct and write a bash script to automate the process for all the genes.
(It has a hacky fix, where the bash script needs to be run again for each line in the file,
for run in {1..48} #change this!
I was too lazy to debug it that day and just wanted to get this finally done).
The source code is provided here, and contains reasonable comments and spits out some debugging info when you run it.
That’s all.
Leave a comment if you have questions 🙂
appendix
Emailed from me to ga4gh-dwg :
With help from biostars*, I finished pulling out the KIR and HLA regions and fixed the SMA region.
The updated collection may be publicly viewed at
https://workbench.qr1hi.
The updated collection README and scripts are at
https://github.com/nouyang-
Of note, I did not mirror the IMGT HLA contents (ftp://ftp.ebi.ac.uk/pub/
Feedback appreciated!
Thanks,
–Nancy
*credit to https://www.biostars.org/p/